Computer vision is already transforming healthcare, medical diagnosis, and drug development. Here are some notable examples:
- The Food and Drug Administration has authorized 882 AI/ML-enabled medical devices as of May 13, 2024. Most are in radiology.
- AlphaFold DB by Google DeepMind provides 200+ million predicted protein structures, used by millions of researchers worldwide.
- A single CNN already achieved dermatologist-level performance on 129,450 images across 2,032 diseases back in 2017.
In short, computer vision has a wide range of use cases in healthcare, from image creation to pathology analysis to diagnosis to outcome prediction.
Computer Vision in Healthcare | Unitlab Annotate
All these innovations have one component in common: high-quality, annotated medical images in their training/testing datasets. In today's post, we will cover medical image labeling from the beginning.
By the end of the post, you will learn:
- What medical image annotation is
- The difference between regular vs. medical image labeling
- HIPAA compliance
- Key annotation techniques and tools
Let's get started.
What is Medical Image Labeling?
In one phrase, labeling medical images. It is the process of annotating medical image data such as X-ray, CT, MRI scans, mammography, ultrasound, and others.
The annotated data is then used to train AI models for medical image analysis, diagnosis, and outcome prediction. These models help doctors detect disease earlier, make more accurate decisions, save time, and improve treatment plans for patients.
The intuition and purpose behind medical and regular photo annotation are the same, but their implementation differs significantly, as we’ll see next.
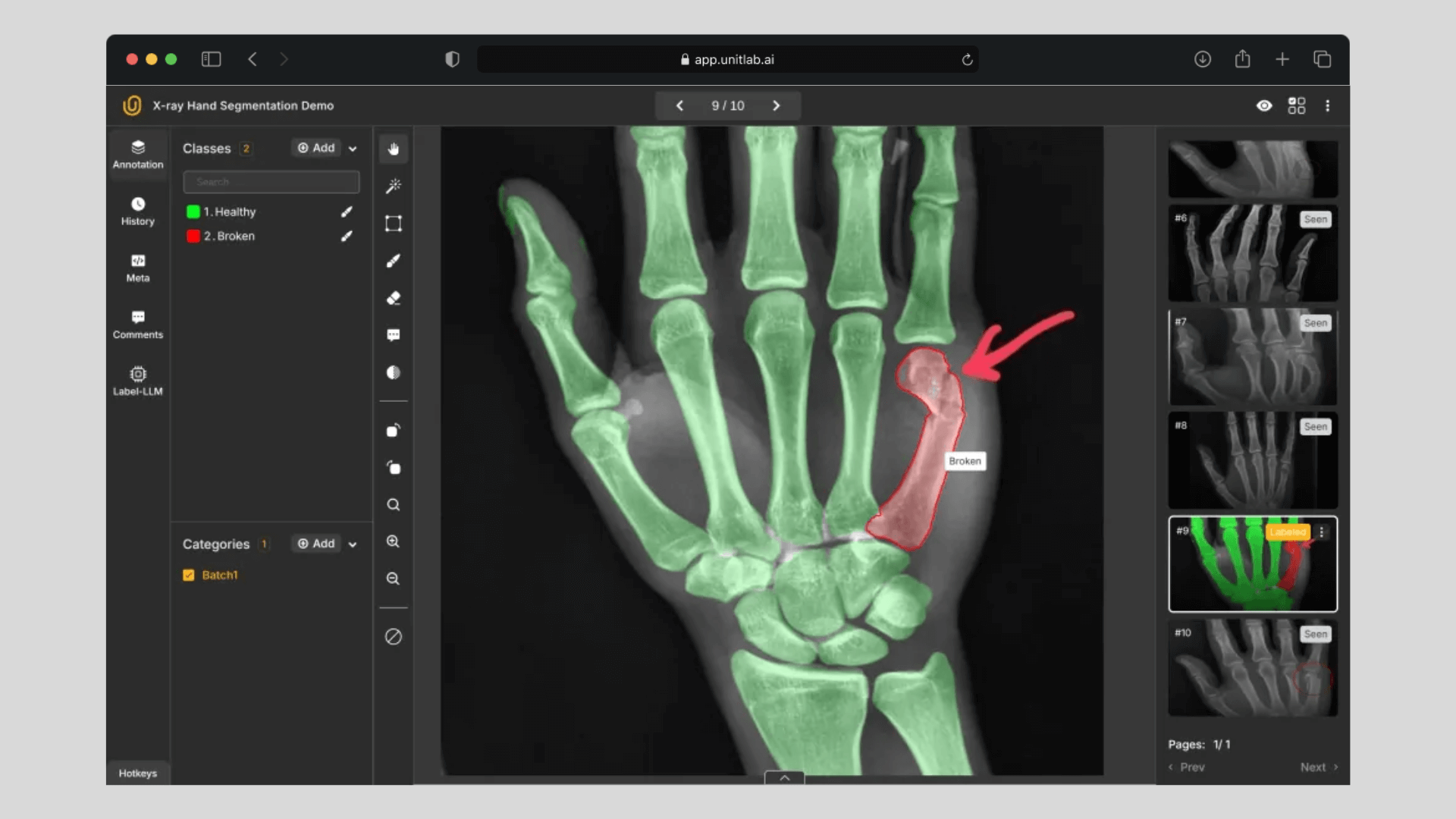
Regular vs. Medical Annotation
Here is the summary table that shows their differences:
| Medical Image Annotation | Regular Image Annotation |
|---|---|
| Protected data by Data Processing Agreements (DPA) | More accessible data |
| DICOM format | JPG, PNG, WEBP, etc |
| Multi-slice annotations | One-slice annotation |
| 16-bit color profile (65,000 values) | 8-bit color (250 values per color) |
| Related measurement units inside files | Relative measurements to the camera |
| Exceptional domain expertise required | Domain expertise not strictly necessary |
| Requires HIPAA compliance | No compliance required |
Data Availability and Collection
First of all, medical data is strictly private, thus requiring very serious treatment. It is subject to many country-specific and international regulations (HIPAA, DPA, FDA, etc.).
Access to medical data archives is reserved only for accredited medical professionals and companies. Handling all the paperwork and infrastructure is time-consuming. Additionally, any newly collected medical data usually requires anonymization, the process of de-identification, further complicating the process.
It’s close to impossible to develop a balanced dataset in medical imaging without long waiting times and deep pockets. Additionally, some diseases are so rare and hard to detect that false positives and negatives are bound to occur.
Most other visual data do not have such a long, complicated collection process. By comparison, their data collection looks very easy and fast.
DICOM Format
Second, most medical imaging is expected to be in DICOM (Digital Imaging and Communications in Medicine). Regular photo data do not need to follow a specific format.
DICOM is the global standard for storing, transmitting, and printing medical images. In a nutshell, DICOM has these features:
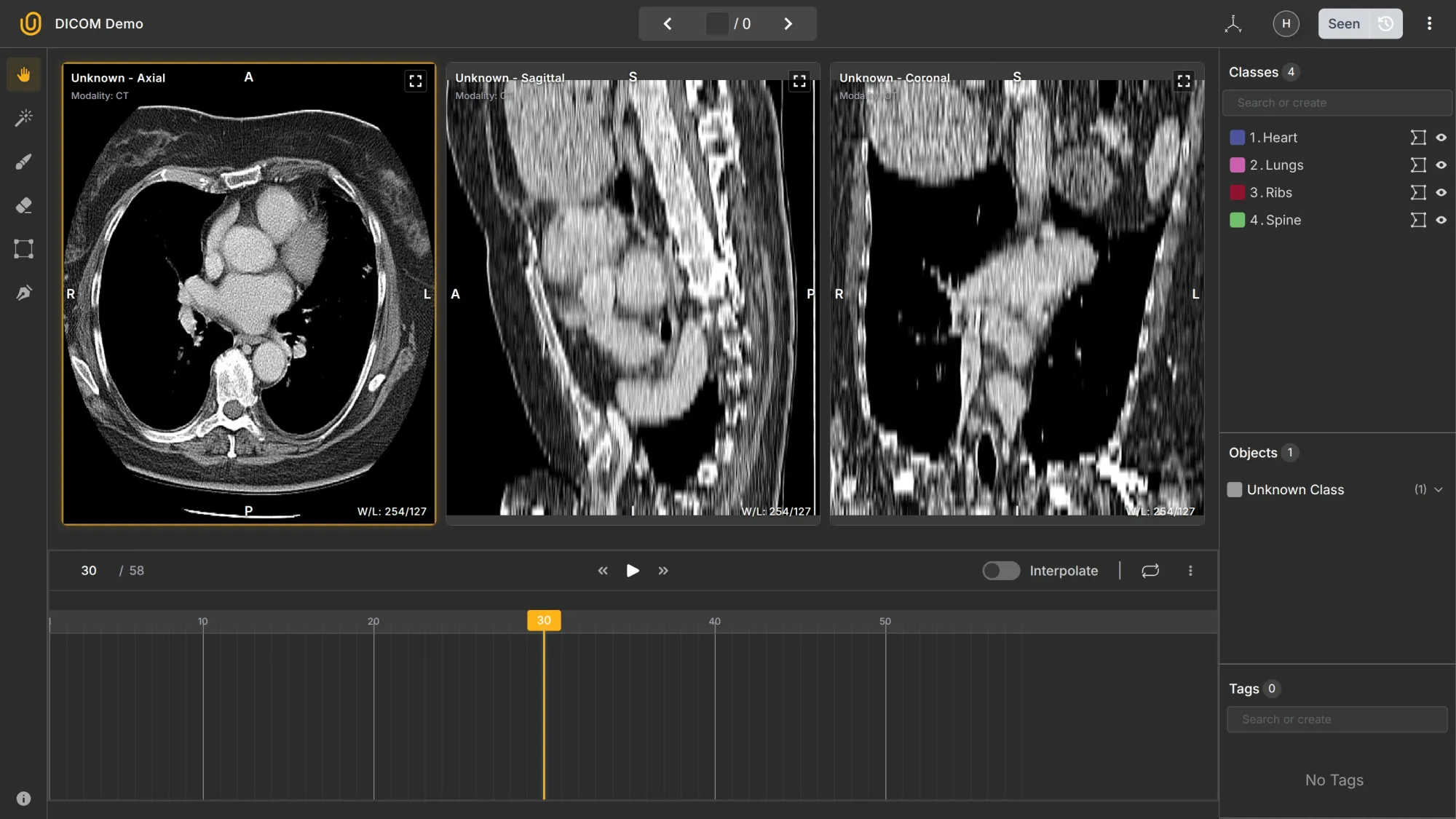
- Multi-slices → A DICOM study can contain dozens or hundreds of consecutive 2D slices of the same body region (e.g., 58 slices through the chest).
- 2D and 3D → Each slice is 2D, but when combined, they form a 3D volume. Annotation tools can reconstruct or navigate through that volume interactively.
- Multi-planes → You can view the same dataset in different planes:
- Axial (top-down)
- Sagittal (side)
- Coronal (front)
Together, these make DICOM data volumetric and spatially consistent; that’s why it’s used for medical imaging rather than formats like PNG or JPEG.
Although the DICOM format is not strictly necessary for training computer vision algorithms, it's highly beneficial to preserve it for data labeling purposes.
Multi-layer, lossless TIFF (Tagged Image File Format) can also be used for storing medical photos. However, it's a wild west out there because of many different variations in the format. Jokingly, TIF is referred to as “Thousands of Incompatible Formats.”
Sometimes, ultra-high-resolution image formats are used by researchers, such as Leica and Apeiro’s SVS. However, these formats typically do not support modifications or annotations.
In comparison, regular visual data have none of these overheads, making them fast, easy, and cheap to store and annotate.
Annotation
Answer this question without googling:
How can an annotator segment a faint, irregular tumor on multi-slice MRI with overlapping tissues and modality-specific contrast?
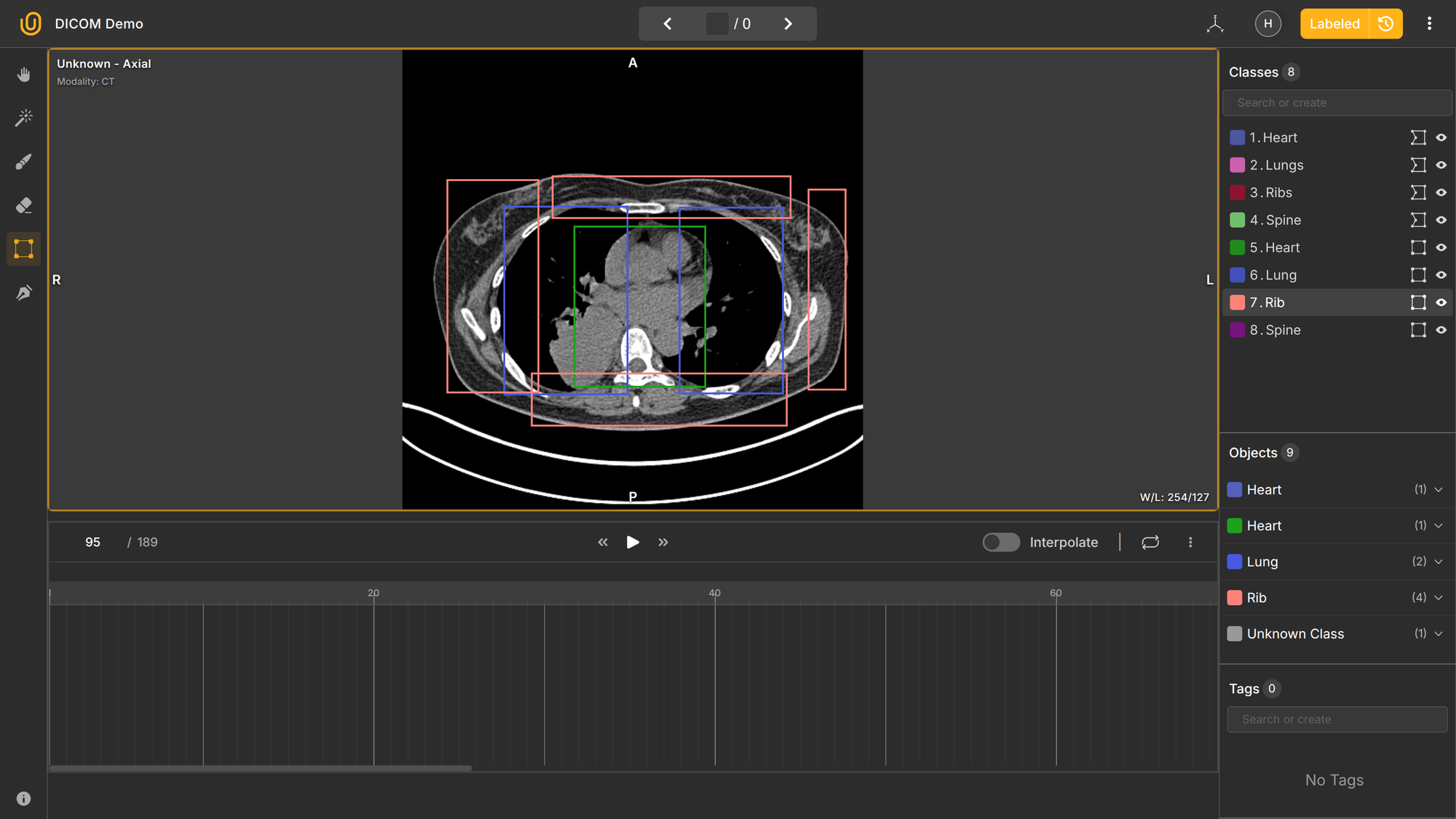
We are certain that 95% of the readers will have no idea what is going on. It is highly probable that most people, even data annotators without medical knowledge, freeze when they see the annotation dashboard for medical labeling:
For starters, what is Axial, Sagittal, and Coronal? What is interpolation in this context? 3D and 2D? Multilayers? 58 frames in the Axial section? Understanding the conventions, let alone annotating medical images, is not an easy task.
Obviously, medical annotation is much different from general data labeling:
Annotate every pedestrian and vehicle with polygons. If an object is occluded, label it as if it were fully visible. Assign new classes called “Shadow” and “Reflection” for shadows and reflections.
With experience and guidance, any sane adult can follow the second instruction and label general objects like cars and pedestrians. The first one? It requires years of rigorous academic training and job experience.
For example, occluded objects refer to objects partly visible in the image. As a rule of thumb, you should label them as if they were fully visible. But what if you can't differentiate them? Are the lower parts of the lungs behind or in front of the diaphragm?
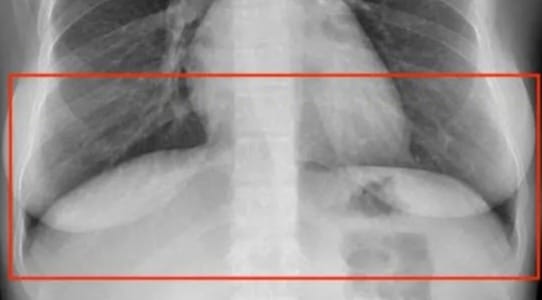
The answer is both.
Medical image annotation is done by domain experts, doctors; rightly so. Even small errors can lead to a patient being diagnosed inaccurately. That’s why we probably won’t see a crowdsourcing platform like Amazon Mechanical Turk for medical data labeling.
HIPAA Compliance
People, companies, and tools that handle sensitive medical information in the USA must follow HIPAA, the Health Insurance Portability and Accountability Act of 1996. HIPAA had a huge impact on other related regulations outside the US as well (GDPR in the EU, for example).
HIPAA is a law that is not to be taken lightly (you may end up paying millions in fines). Every part of medical data storage, collection, annotation, and usage should strictly follow its guidelines.
For instance, clinical image datasets must include a clear record of who contributed to each annotation. Regulatory approval requires proof of annotation authorship, dataset integrity, and detailed review history.
Both the U.S. Food and Drug Administration and the European Commission set expectations for how datasets should be structured when used to build diagnostic models.
HIPAA includes many additional points for other workflows in medical image annotation, such as data access:
- Does the facility where the annotators label the data have means of access control?
- Do the annotators with access to electronic patient health information (ePHI) access their mobile devices while working?
- Do you maintain a detailed inventory of all hardware involved in this project and a record of its movement?
- Are human annotators trained for, and aware of, these requirements?
A good HIPAA checklist can be found here.
All the factors above make medical image annotation fundamentally different and extremely challenging compared to ordinary visual data labeling.
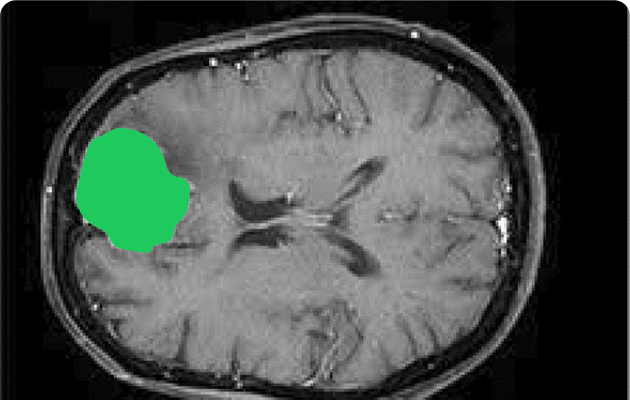
Key Annotation Techniques
Depending on the type of organ, bone, or cell being annotated, image annotation types likely differ:
- Bounding boxes: Identify regions of interest, like tumors or organs. Used in radiology (tumor localization), pathology (cell detection), dermatology (lesion detection).
- Polygon and brush tools: Capture complex shapes, such as lung lobes or lesions. Usually for pulmonology (lung segmentation), oncology (tumor margins), cardiology (heart chambers).
- Semantic segmentation: Classify each pixel to map organs and abnormalities. For radiology (organ and tumor segmentation), neurology (brain structure mapping), ophthalmology (retinal layer analysis).
- Keypoint annotation: Mark critical points in anatomical structures. Use cases in orthopedics (joint landmarks), surgery (instrument tracking), ophthalmology (optic disc and fovea points).
- 3D annotation: Label volumetric scans from MRI or CT for higher precision. Used in neurosurgery (brain tumor planning), radiology (multi-slice CT/MRI), cardiology (vessel and chamber analysis).
Although most medical image annotation is done manually by experienced researchers and doctors, auto-annotation tools can be of much help. They can speed up labeling data tremendously.
Some platforms also provide interpolation: automatic labeling between two slices. For example, you annotate slides 30 and 35, and the software tries to auto-annotate slices 31-34. It doesn't always work accurately, but can be a good side help.
In the video below, we annotated only one slice, and let the machine annotate the rest based on that. Generally, the smaller the range, and the more the annotations, the better the interpolation works.
Tools and Technologies
Default DICOM viewers can be used to handle basic annotation, such as bounding boxes, arrows, or polygons. However, their data format is not usually suitable for ML workflows. They lack identifiable IDs, labeling queues, and other attributes for deep learning frameworks such as PyTorch and TensorFlow.
Therefore, professional and open-source data annotation platforms have evolved to produce formats for these medical images. They are used for training ML algorithms.
The peculiar feature of medical image annotation is that, in addition to standard criteria for data annotation platforms, you need to take into account the nature and requirements of medical imaging:
- Is DICOM supported?
- Is it HIPAA compliant?
- Does it provide labeling services for medical imaging?
- What annotation types does it have?
- Can you export data to deep learning frameworks like Pytorch?
Some of the prominent data annotation platforms include V7 Labs, Label Studio, and SuperAnnotate. There also exist open-source DICOM-compatible tools, but you should ensure that you are following HIPAA requirements yourself.
Conclusion
Computer vision is transforming healthcare, no question. It relies on high-quality, balanced medical image sets. Medical image annotation is the process of tagging medical images so that deep learning frameworks can process them.
However, there are quite a few significant challenges: regulations, insufficient data, domain requirements, and technical difficulties. A HIPAA-compliant data platform can handle much of the headache.
Still, medical image annotation is the backbone of medical AI and continues to drive future advancements in human knowledge and ingenuity.
Explore More
Check out these articles for more on annotating medical photos:
- Computer Vision in Healthcare: Applications, Benefits, and Challenges
- Guide to Pixel-perfect Image Labeling
- Why Outsource Medical Data Labeling?
References
- Alberto Rizzoli (Jan 31, 2022). The Ultimate Guide to Medical Image Annotation. V7 Labs: Source
- Cogito Tech (Jul 10, 2025). Medical Image Annotation and Labeling Services: A Complete Guide 2025. Cogito Tech: Source
- Fabio Galbusera and Andrea Cina (Feb 06, 2024). Image annotation and curation in radiology: an overview for machine learning practitioners. European Radiology Experimental: Source
- iMerit (no date). A Complete Guide to Medical Image Annotation. iMerit: Source

![The Guide to Medical Image Annotation: Essentials, Techniques, Tools [2025]](/content/images/size/w2000/2025/12/medical.png)